Prof. Patil Selected as Faculty Co-Director of the University of Utah’s DATASET Initiative
Associate Prof. Sameer Patil from the Kahlert School of Computing has been selected as a faculty co-director of the Data Science & Ethics of Technology (DATASET) Initiative at the University of Utah to serve alongside co-director Prof. Manish Parashar. Prof. Patil replaces past co-director Prof. Olivia Sheng who is leaving the University of Utah this summer to join the W. P. Carey School of Business at Arizona State University.

The DATASET Initiative is a part of the One Utah Data Science Hub, a university-wide effort designed to enhance research and infrastructure in data science and data-enabled science. DATASET was launched in the fall of 2022 to engage in foundational questions about the role of data in science and society. The initiative aims to bring together research and expertise in the theoretical, technical, ethical, historical, and policy/legal dimensions of data across the University of Utah. The goals of the DATASET initiative are well-aligned with Prof. Patil's courses on Ethics in Data Science and Human Aspects of Security and Privacy. “I am honored to have been selected for the role of co-director and excited to leverage the position for promoting data sharing and open science to facilitate transparency, verifiability, and replication of data-driven research results,” Patil said.
In his role as co-director, Prof. Patil will help develop the vision and specific activities of the DATASET initiative and engage with the DATASET Advisory Board. Associate Director of the One Utah Data Science Hub Penny Atkins said, "We were impressed by Prof. Patil's experiences, perspectives, and goals related to DATASET and believe that he will be an asset to our initiative going forward."
Kahlert School Research on Visualizations for Metabolic Networks Published in Nature Cell Biology and Science
In September 2019, Jordan Berg, then a PhD student in the lab of Professor Jared Rutter in the Biochemistry Department, contacted Professor Bei Wang Phillips in the Kahlert School of Computing and the Scientific Computing and Imaging Institute regarding how best to design a tool for the analysis and visualization of metabolic networks. Over the next four years, then PhD student Youjia Zhou from Professor Wang Phillips’ lab collaborated with Jordan and Professor Rutter’s team. The collaboration has contributed toward two recent prestigious publications: one in Nature Cell Biology led by Jordan (Jordan A. Berg et al., 2023) and another in Science led by Dr. Kevin G. Hicks (Kevin G. Hicks et al., 2023). Berg and Zhou defended their PhDs in the Springs of 2022 and 2023, respectively. Dr. Zhou’s PhD thesis is titled “Topology-Based Visualization of Graphs and Hypergraphs.”
Metabolism is an important process in our bodies that affects various aspects of our health, such as cell growth and development, stress response, and energy production. It is a complex, interdependent network, and small changes in one area can have widespread effects throughout the entire metabolic system. Existing tools for analyzing metabolic networks suffer from various problems: they may be outdated, have limited functionality, be too technical to run for a biochemist, or completely unusable. To address these shortcomings, Zhou and Wang Phillips collaborated with Berg and Rutter’s team to develop new tools that leverage interactive graph visualizations. These tools are designed to analyze complex metabolic networks with a focus on detecting patterns and studying interactions between proteins and metabolites.


“Our vision is to develop practical visualizations of networks that help domain scientists with their knowledge discovery process,” said Professor Wang Phillips.
Pattern Recognition
The first interactive tool is called Metaboverse, a user-friendly tool that helps with the exploration of metabolic networks. In particular, it enables pattern recognition from a large, varied library of possible metabolic patterns. In the context of metabolism, a reaction pattern refers to a metabolic reaction that shows some change between the inputs and outputs of the reaction. Metaboverse enhances the understanding of detected patterns by providing a dynamic exploratory interface, which helps domain scientists come up with new ideas.
Previously, scientists searched for desired patterns manually, which was time-consuming and incomplete. Similarly, other available tools could find and identify only a few types of patterns and had the potential to overlook crucial insights. Metaboverse helps solve these problems: it introduces a broad collection of patterns and, using automated techniques, provides a quick and accurate way for scientists to detect complex patterns that they might have previously missed.
In terms of practical applications, Metaboverse was used to find patterns in the metabolism of early-stage human lung adenocarcinomas (LUAD), a type of lung cancer. The goal was to identify which metabolites could be used to diagnose the disease at an early stage. Metaboverse was able to prioritize patterns in nucleotide metabolism reliably; consistent with the original study as well as the manual re-analysis of the data using other existing tools. In addition, Metaboverse helped discover new patterns related to xanthine metabolism and a group of reactions linked to a decrease in lysine. These discoveries were made using data from a previous study where changes in these metabolites were observed but not fully explored.
Analysis of Protein-Metabolite Interactions
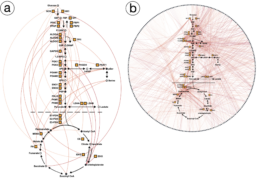
Protein-metabolite Interactions (PMIs) indicate how proteins and metabolites communicate metabolic status to different cellular processes. In metabolic networks, finding PMIs that mediate these networks can be difficult because they often have low affinity. To help solve this problem, Zhou and Wang Phillips worked with Berg and Rutter’s team to create MIDAS, a library that helps find PMIs systematically. Specifically, the researchers helped develop Electrum, a flexible, user-friendly visualization portal that aids in the understanding and analysis of the PMI data generated by MIDAS. While other general visualization platforms are unable to handle PMI data because of the specific analytical capabilities it requires, Electrum can differentiate between known and unknown PMIs, which can lead to new discoveries.
The usefulness of Electrum was demonstrated through a case study in which interactions within and between pathways in carbohydrate metabolism were discovered. Carbohydrate metabolism enzymes drive most of the energy production and biosynthetic precursor generation of cells, and their regulation involves metabolite interactions. The MIDAS platform identified 830 putative PMIs, many of which were previously unknown. Electrum provided interactive visualizations to help domain scientists comprehend the PMIs within and between pathways, and the visual analytics results revealed regulation within and between pathways in carbohydrate metabolism. The collaboration led to a recent publication in Science led by Dr. Kevin G. Hicks.
In the future, the researchers will continue developing and enhancing interactive visualization tools to help domain scientists better explore and understand complex networks.
Award-winning Research from the Kahlert School Examines Student Understanding of Code Quality
Kahlert School of Computing Ph.D. student Sara Nurollahian and Assistant Prof. Eliane Wiese have received the best paper award for the Software Engineering Education and Training Track at the International Conference on Software Engineering (ICSE 2023) for their paper titled Improving Assessment of Programming Pattern Knowledge through Code Editing and Revision that was presented at the conference earlier in May 2023.
When programmers write code, it’s not enough for a program to run, or even to work, as expected. Even if a computer is capable of parsing and properly executing the code, humans need to be able to read and understand it as well in order to maintain, test, and debug it over the course of its life. Given the complex nature of modern code, how can computer science students learn to think about, design, and write quality code that is highly technical and highly readable? Addressing this larger question is the motivation behind the research in the award-winning paper. Advised by Prof. Eliane Wiese, Nurollahian examined how student coders demonstrate understanding of their code through different activities. “ICSE is the premier conference on software engineering and it’s exciting to have Sara present her work in their education track, which reaches a broad range of Computer Science education researchers around the world,” Prof. Eliane Wiese said.


In their research, Nurollahian and Wiese used a survey to measure student understanding of two important code quality issues: avoiding repetitions and returning boolean expressions rather than literals in simple methods. These types of quality issues are similar to simplifying math expressions. For example, it is difficult to think of the two integer values for x and y for the equation 4x + 8y + 3 = 39, but it is a lot easier for this equivalent one: x + 2y = 9.
A common approach to research in this area is to measure student' understanding by examining the code they write. However, students may write elegant code without understanding the principles behind it or write poor-quality code even if they know better. Instead of looking only at code writing, the study additionally asked students to revise their own code and edit some code written by others. For code writing, students were given the signature of a method along with a description of its desired behavior and asked to fill in the body. For editing, students were given low-quality code blocks written by others and asked to improve them. Similarly, for revising, students who wrote low-quality code in the writing task were shown their own code and asked to improve it. However, for revising, students were given a series of hints, starting with a simple prompt to revise (without indicating what was wrong or how to fix it), and ending with an example for a similar code block.
“These three tasks allowed us to examine the revising and editing performance of students who wrote low-quality code, considering that students whose code is low in quality due to knowledge gaps will not be able to perform well in revising and editing without receiving extra support, ” Nurollahian said. Students who wrote poor-quality code needed different levels of hints to revise successfully, allowing the researchers to differentiate between knowledge gaps and other factors, such as motivation. For example, over half of the students who returned boolean literals in the writing task were able to revise successfully when prompted to improve (without getting information on what to fix or how), suggesting that they simply needed more motivation. However, the majority of the students who wrote code that contained repetitions were not able to revise successfully, even after seeing hints on what to change and an example of implementing the change in a similar code block, indicating that they needed greater knowledge in this area. The editing tasks additionally showed that code writing can give an incomplete picture of student understanding. While students who wrote well initially were generally able to edit more successfully, there were exceptions in both directions. For returning boolean literals (vs. expressions), between one quarter and one third of students who wrote poorly were successful in editing someone else's code, while a similar percentage of students who initially wrote well made unsuccessful edits. For code repetitions, fewer students overall wrote or edited successfully, but the general pattern still held.
These findings highlight the benefit of using a variety of tasks to measure student understanding of code quality. The work also underscores the importance of further research to examine the quality issues that are challenging for students. As Nurollahian explained, “The distinction between simple and complex quality issues can help instructors allocate their time and effort to the more challenging ones. Additionally, distinguishing between students who have deep conceptual gaps and those whose low-quality code is due to other reasons, such as lack of motivation, can help appropriately tailor instruction, feedback, and learning interventions.”
A New Technique by Kahlert School Researchers Paves the Way for Enhancing Training Efficiency for Physics-Informed Neural Networks
Kahlert School of Computing researchers Ramansh Sharma, an incoming PhD student, and Dr. Varun Shankar, an Assistant Professor, have developed a novel method that expedites the training of a specific type of artificial intelligence (AI) system known as Physics-Informed Neural Networks (PINNs). This new technique, referred to as Discretely-Trained PINNs (DT-PINNs), offers promising improvements in training speed and efficiency and has the potential to broaden the applications of PINNs while lowering cost. The work was published at the Conference on Neural Information Processing Systems (NeurIPS) 2022. NeurIPS is a key top-tier event for those in the artificial intelligence field, showcasing research that spans machine learning, computational neuroscience, and more.
Neural networks form the foundation of modern AI technology. Inspired by the human brain, they are comprised by interconnected layers of nodes, or “neurons,” that collaborate to learn from data and make predictions or decisions. PINNs are a unique type of neural network that incorporate principles of physical laws into their structure. PINNs are used in a myriad of fields, including engineering, physics, and meteorology, where they can, for example, help predict structural responses to stress or forecast weather patterns.


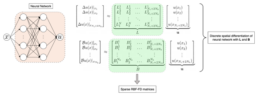
Training neural networks is a complex and resource-intensive process. The system needs to adapt the strength of connections between nodes based on input data, a task involving the calculation of many partial derivatives. This is especially challenging for PINNs since the incorporation of exact derivative terms into the training process makes these connections more time-consuming to compute. The DT-PINN method developed by Sharma and Shankar addresses this issue by replacing these exact spatial derivatives (which define how a quantity is changing in space) with highly-accurate approximations calculated using a technique called meshless Radial Basis Function-Finite Differences (RBF-FD). Despite the fact that neural networks are traditionally trained using 32-bit floating point (fp32) operations for the purposes of speed, the University of Utah researchers found that using 64-bit floating point operations (fp64) on the Graphics Processing Unit (GPU) for DT-PINNs leads to faster training times.
“Our work on DT-PINNs represents a substantial progression in the training of PINNs. By improving the training process of PINNs, we have expanded their potential applications and reducing computational costs," Prof. Shankar said. Sharma and Shankar demonstrated the efficiency of DT-PINNs through several experiments, showing that they could train 2-4 times faster on consumer GPUs without compromising on accuracy compared to training via traditional methods. The researchers applied the technique to linear as well as nonlinear spatial problems and demonstrated its applicability to a problem involving space as well as time dimensions.
The results of the research present a significant stride toward more efficient utilization of PINNs across various scientific and technological fields. By reducing the time it takes to train these networks, researchers and practitioners can quickly develop and deploy sophisticated models, aiding our understanding and prediction of complex physical phenomena.